eQuilibrator: Redox Reactions
An electron carrier is a molecule that can stably occupy reduced and oxidized states - like NAD+ and NADH diagrammed below. Most biological redox reactions involve the enzyme-catalyzed transfer of electrons to or from an electron carrier. If you took a biochemistry class at some point you probably learned about the common biological e- carriers: NAD(P)H, FADH2, various quinones, ferredoxins and, if you were lucky, the archaeal carrier coenzyme F420.
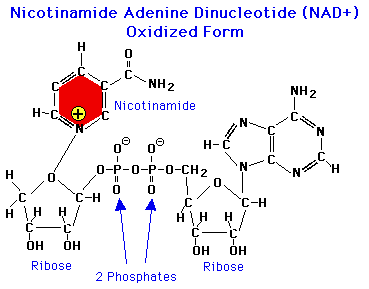
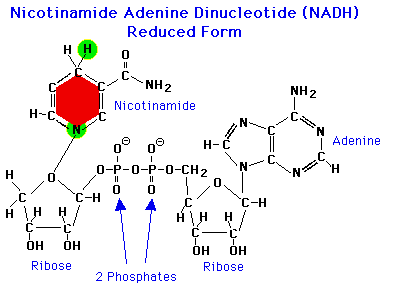
Electron carriers are characterized quantitatively by their reduction potentials (usually in mV units). A carrier with a +100 mV potential will more readily accept electrons than one with -100 mV potential because electrons flow to positive potential by convention. If you know the potential of the electron donor and acceptor in a redox reaction, you can calculate the ΔrG of that reaction (read more here and here).
Many biological electron carriers are straightforward to think about in that they are small, defined molecules that we can purify and characterize in the lab. NAD(P)H and FADH2 are great examples - it is relatively easy to measure the redox potential of these carriers because it is clear what they are. But this is not the case for all e- carriers. Ferredoxins, for example are proteins containing an iron-sulfur cluster and their redox potentials can vary by more than 200 mV depending on the exact sequence of the protein. As such, we can’t reasonably attach a number to ferredoxin in eQuilibrator - unless you have a list of potentials for all the ferredoxin genes in nature you want to share.
Iron is a problem in general, really. Some bacteria draw electrons (and, thereby energy for growth) from iron-containing minerals. But these minerals can differ in composition and, therefore, redox potential pretty substantially. On top of that I am not a minerologist. So Elad came up with an elegant solution to dealing with redox reactions involving problematic donors or acceptors in eQuilibrator - you just tell us what the potential is.
You can try it by example using the oxidation of glucose to gluconate - “glucose + H2O = gluconate”. If the electrons will be donated to the Spirulina ferredoxin II, which has a -305 mV potential you can simply enter -305 into the text box labeled “e- potential” and click “Update” to calculate the ΔrG’ value you came for.