DABs are inorganic carbon pumps
This post is a reflection on and summary of my recent work with Jack Desmarais and several other members of the Savage lab: Desmarais JJ, Flamholz AI, Blikstad C, Dugan EJ, Laughlin TG, Oltrogge LM, Chen AW, Wetmore K, Diamond S, Wang JY, Savage DF. 2019. DABs are inorganic carbon pumps found throughout prokaryotic phyla. Nat Microbiol 4:2204–2215. doi:10.1038/s41564-019-0520-8
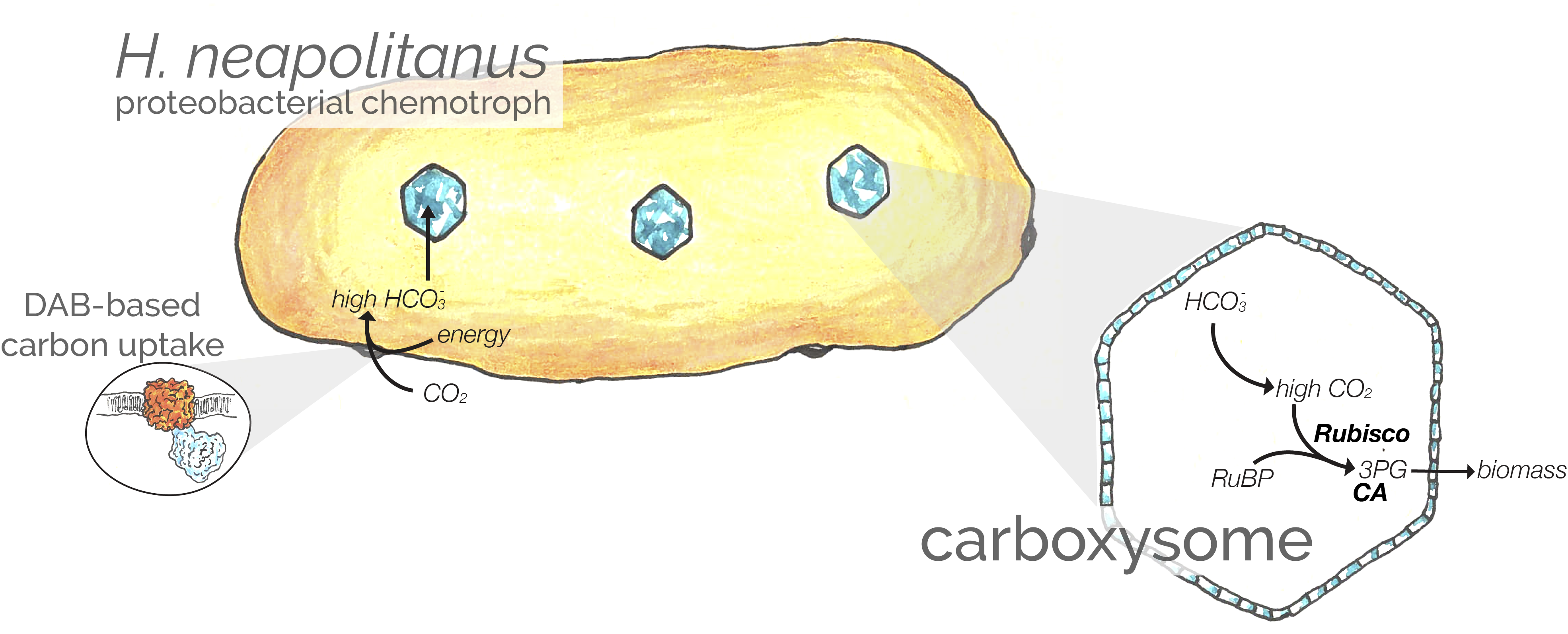
When I started my PhD studying the carboxysome-based CO2-concentrating mechanism (CCM) found in bacteria, it was unclear if we knew all the genes making up this exquisite system. Based on the accumulated data, we thought there were about 15 genes - roughly 10 genes making up the carboxysome structure and 2-5 genes for inorganic carbon transport. Also, when we started, the transporters in the proteobacterial family had not yet been found. So my goal was to determine (i) whether other genes are required for the CCM and (ii) which genes perform the essential transport activity. We were happy to discover that the Arkin and Deutschbauer labs upstairs had developed a clever extension of TN-Seq called RB-TNSeq, which greatly simplifies whole genome knockout screens in bacteria. When Jack Desmarais joined the lab, we decided to focus on the model proteobacterium, H. neapolitanus because (i) we can purify its carboxysomes, and (ii) earlier studies had focused on cyanobacteria and so less was known about the proteobacterial CCMs.
Using RB-TNSeq we compared the phenotypes of 70,000 knockout mutants between ambient air - where the CCM is required for growth - and elevated CO2 (where the CCM is dispensable). 17 genes appear to be required for CCM function, including all known carboxysome genes, three transcriptional regulators, a putative Rubisco chaperone, and a pair of candidate transporters. Building on three recent papers from Kathleen Scott, we showed that a pair of genes - DabAB2 - are both necessary and sufficient for inorganic carbon uptake in our E. coli reporter strain. Jack also showed that DabAB2 form a zinc-containing complex, that the putative zinc-binding resiudes are required, and that the uptake substrate if probably CO2 (not HCO3-). Altogether we think the data imply pretty strongly that DABs are two-protein complexes that convert extracellular CO2 into intracellular HCO3- using an energy-coupled zinc-based carbonic anhydrase-like mechanism. We think the energy source is an H+ or Na+ gradient.
In homage to the open source tradition, we cheekily call these transport complexes DABs for “DABs accumulate bicarbonate.” The illustration by Rachel Shipps above gives a rough sense of what we think this complex looks like, though we can only speculate about how it works. Which we do, of course, in the paper. Thanks to Rachel Shipps for her amazing illustrations, Eli Dugan for making so many useful E. coli mutants in a pinch, Kelly Wetmore and Morgan Price for much help with RB-TNSeq methods and analysis, Spencer Diamond for the massive upgrade in phylogenetic analysis, and everyone in the Savage lab who pitched in, especially Cissi, Tom, Luke and Allen. If you are hungry for more detail, you can read my thoughts on the preprint here and Jack’s blog post on the paper over at the Nature Community blogs.