Functional reconstitution of a bacterial CO2 concentrating mechanism
This post is a reflection on and summary of my recently published work with collaborators: Flamholz AI, Dugan E, Blikstad C, Gleizer S, Ben-Nissan R, Amram S, Antonovsky N, Ravishankar S, Noor E, Bar-Even A, Milo R, Savage D. 2020. Functional reconstitution of a bacterial CO2 concentrating mechanism in E. coli. Elife 9. doi:10.7554/eLife.59882
All plants, algae and cyanobacteria rely on the Calvin-Benson-Bassham (CBB) cycle for growth. Rubisco is the central enzyme of the CBB cycle and the most abundant enzyme on the planet: it does the tricky bit where CO2 gets “fixed” onto a soluble sugar. While it is often said that rubisco is “slow,” it is actually an average enzyme in terms of “turnover number” (maximum rate per active site, kcat). The true rate of rubisco carboxylation is much slower than its kcat, however, because rubisco can react non-specifically with O2. I certainly think it’s surprising that rubisco is not faster or more specific given how important and abundant it is.
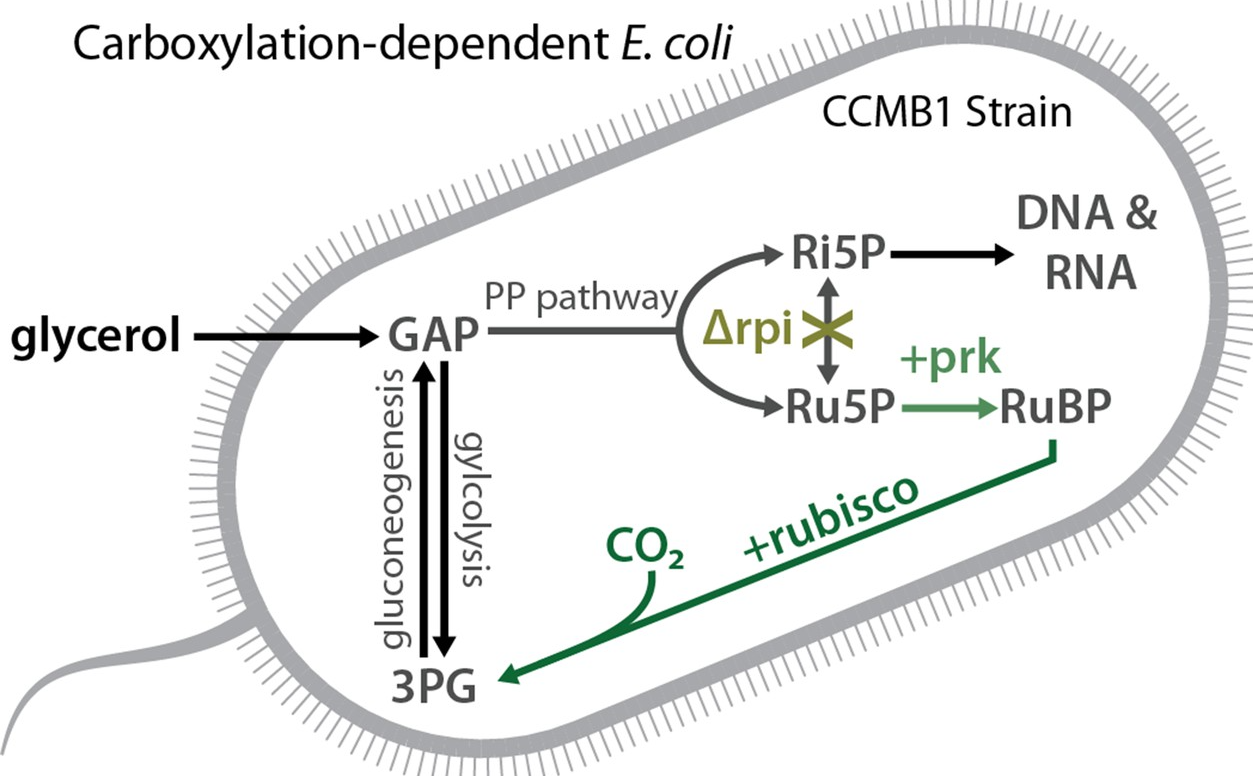
Many photosynthetic organisms have evolved CO2-concentrating mechanisms (CCMs) that compensate for the relative inefficiencies of rubisco by elevating CO2 levels near the enzyme. Elevated CO2 is doubly beneficial, bringing rubisco closer to CO2-saturation and also excluding O2 from the active site (as explained here). CCMs are very significant on the global scale, responsible for perhaps 50% of global photosynthesis. CCMs are also very significant to the organisms harboring them: CCM mutants typically entirely fail to grow in ambient levels of CO2 (0.04%) and require CO2 supplementation to enable robust growth (usually 1-5%).
In this paper we paid homage to Richard Feynman’s famous quote “What I cannot create I do not understand” building a bacterial CO2-concentrating mechanism from its constituent parts in a non-native host, namely E. coli. To do this, I first designed an E. coli strain called CCMB1 (diagrammed above) whose growth in defined media depends on rubisco expression. This strain grows only when rubisco is expressed, and even then only when high CO2 levels are provided in the growth chamber. By expressing the known components of the bacterial CCM, I showed that a functional CCM permits CCMB1 E. coli to grow in ambient CO2 levels. Via genetics experiments I showed that this growth requires all the known components of the CCM and none of the putative auxiliary genes. Furthermore, using isotope tracers, I showed that CCMB1 does in fact use rubisco to fix CO2 into the molecules making up its biomass.
As a result of this confirmatory data, I can confidently say that this represents the first functional reconstitution of any CCM. Reconstitution systems are be very useful for testing hypotheses about genes and proteins, e.g. “is this gene required” or “how does this mutant affect the function of the CCM.” Along these lines, I used CCMB1 to show that rubisco chaperone genes (cbbOQ and acRAF) are not strictly required for the CCM to function. Many researchers are interested in using these types of experiments to delineate a “minimal” CCM construct for expression in crop plants because many crops lack CCMs (e.g. soy, wheat) and modeling suggests that crop yields could be improved by increasing the efficiency of CO2 fixation. Indeed, various outlets covering of our paper (eLife, Nature, IGI, F1000) highlight these motivations. I am glad for the coverage, but, as I have written elsewhere, I am not sure that these efforts will ultimately succeed for the simple reason that plants and bacteria grow very differently. The bacterial CCM might not be compatible with plant physiology. Here, again, CCMB1 could come to the rescue - one could use CCMB1 to test how the functioning of a CCM depends on the growth conditions (e.g. temperature, mixing, surface-to-volume ratio) and infer whether the laborious work of expressing ≈20 genes in a plant is worthwhile.
I’m very proud of this work. It was borne out of my time thinking about CO2 fixation with Elad Noor and Arren Bar-Even in Ron Milo’s lab and I carried the project with me into my PhD in Dave Savage’s lab at UC Berkeley. Finishing this project took me the better part of seven years, years which had a huge impact on my life. In particular, I need to thank my technician and friend Eli Dugan who helped so much with this project and who is about to start on his own PhD journey. I can’t wait to see what he accomplishes.